ZFLNCG00133
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 58.09 |
| SRR1648856 | brain | normal | 41.04 |
| SRR1648854 | brain | normal | 13.65 |
| SRR372802 | 5 dpf | normal | 8.31 |
| SRR1648855 | brain | normal | 8.16 |
| SRR1565805 | brain | normal | 7.90 |
| SRR1565811 | brain | normal | 7.53 |
| SRR1167757 | embryo | mpeg1 morpholino and bacterial infection | 7.48 |
| SRR1565813 | brain | normal | 7.41 |
| SRR1565819 | brain | normal | 7.39 |
Express in tissues
Correlated coding gene
Download
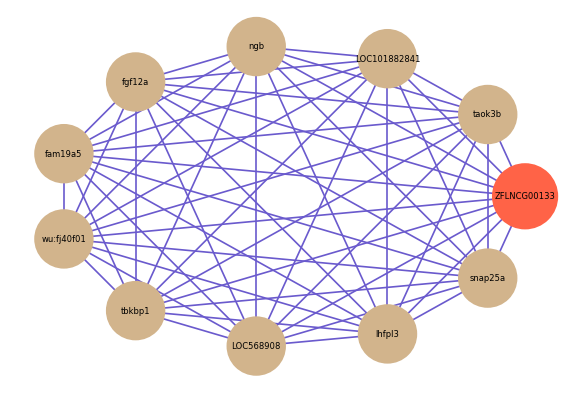
| gene | correlation coefficent |
|---|---|
| taok3b | 0.87 |
| LOC101882841 | 0.86 |
| ngb | 0.86 |
| fgf12a | 0.86 |
| fam19a5 | 0.86 |
| wu:fj40f01 | 0.86 |
| tbkbp1 | 0.86 |
| LOC568908 | 0.85 |
| lhfpl3 | 0.85 |
| snap25a | 0.85 |
Gene Ontology
Download
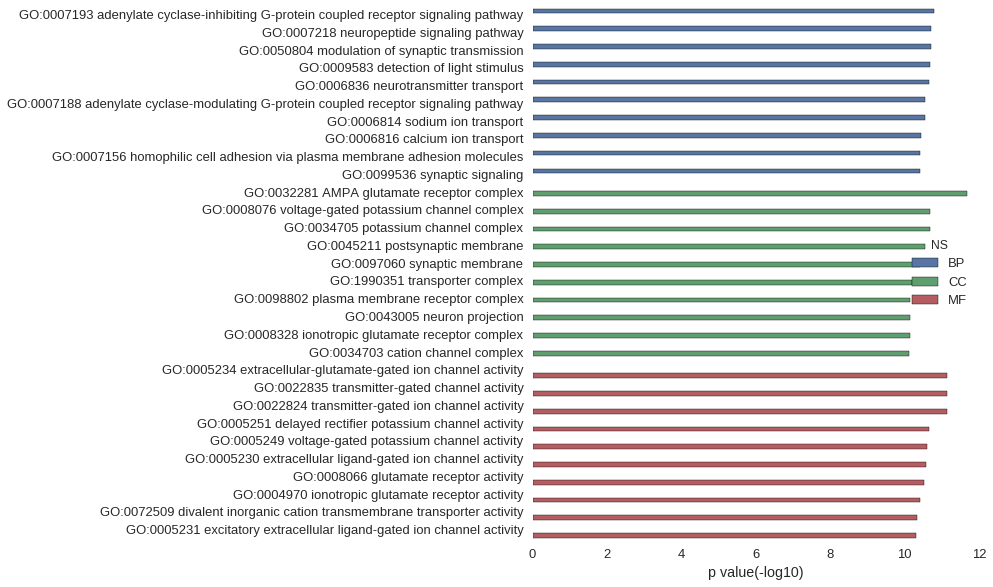
| GO | P value |
|---|---|
| GO:0007193 | 1.61e-11 |
| GO:0007218 | 1.92e-11 |
| GO:0050804 | 1.95e-11 |
| GO:0009583 | 2.05e-11 |
| GO:0006836 | 2.25e-11 |
| GO:0007188 | 2.75e-11 |
| GO:0006814 | 2.75e-11 |
| GO:0006816 | 3.52e-11 |
| GO:0007156 | 3.85e-11 |
| GO:0099536 | 3.93e-11 |
| GO:0032281 | 2.15e-12 |
| GO:0008076 | 2.05e-11 |
| GO:0034705 | 2.05e-11 |
| GO:0045211 | 2.80e-11 |
| GO:0097060 | 3.89e-11 |
| GO:1990351 | 6.31e-11 |
| GO:0098802 | 7.13e-11 |
| GO:0043005 | 7.13e-11 |
| GO:0008328 | 7.28e-11 |
| GO:0034703 | 7.79e-11 |
| GO:0005234 | 7.23e-12 |
| GO:0022835 | 7.32e-12 |
| GO:0022824 | 7.32e-12 |
| GO:0005251 | 2.24e-11 |
| GO:0005249 | 2.49e-11 |
| GO:0005230 | 2.64e-11 |
| GO:0008066 | 2.96e-11 |
| GO:0004970 | 3.90e-11 |
| GO:0072509 | 4.73e-11 |
| GO:0005231 | 4.85e-11 |
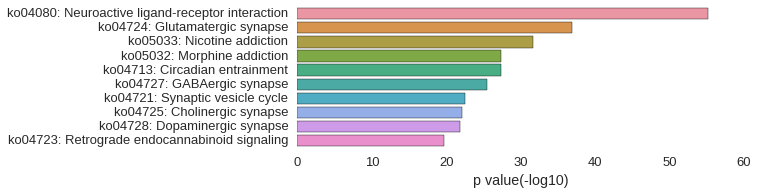
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00133
AGGGTTATTTAGCTTTATTGAATCTGCATATTCGGCATTCCCTTGAGTTCAGTTTCTGGCTCTCGAAACT
ACTTTTTTTTAAATCTTCGTCGTAAGTTCACACATTTCCAAGGAAAAGTAGAAACTAGTAAGAATCATAC
AAGAGGAGCATGTATTTCATTCAAGTGTTTACATAAGCATTTACATTGCCTTGCCAAAAGTAATCAGGCC
TTGACCAATTCTGTCATGTTACTGGATTACACAGTCCCACACTGCAGTTTGTGTTCTGTTATGTTGCATT
TCAAAGCCCCGAATCTAATTTATTAAAACAACATTGTTTTGACATTCCGCACATTTTAATATTTCTCAAA
TACTGCTATTTTTGTTGGATTTGCAGCACATCCGAAAAGTATTCATAGCGCTTCAC
AGGGTTATTTAGCTTTATTGAATCTGCATATTCGGCATTCCCTTGAGTTCAGTTTCTGGCTCTCGAAACT
ACTTTTTTTTAAATCTTCGTCGTAAGTTCACACATTTCCAAGGAAAAGTAGAAACTAGTAAGAATCATAC
AAGAGGAGCATGTATTTCATTCAAGTGTTTACATAAGCATTTACATTGCCTTGCCAAAAGTAATCAGGCC
TTGACCAATTCTGTCATGTTACTGGATTACACAGTCCCACACTGCAGTTTGTGTTCTGTTATGTTGCATT
TCAAAGCCCCGAATCTAATTTATTAAAACAACATTGTTTTGACATTCCGCACATTTTAATATTTCTCAAA
TACTGCTATTTTTGTTGGATTTGCAGCACATCCGAAAAGTATTCATAGCGCTTCAC