ZFLNCG00717
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700534 | heart | control morpholino | 16.81 |
| SRR700537 | heart | Gata4 morpholino | 11.90 |
| SRR1562533 | testis | normal | 1.51 |
| SRR516121 | skin | male and 3.5 year | 1.27 |
| SRR1647681 | head kidney | SVCV treatment | 1.20 |
| SRR1647680 | head kidney | SVCV treatment | 1.16 |
| SRR1647679 | head kidney | normal | 0.97 |
| SRR1205166 | 5dpf | transgenic sqET20 and neomycin treated 3h | 0.92 |
| SRR1647682 | spleen | normal | 0.88 |
| SRR1205154 | 5dpf | transgenic sqET20 | 0.75 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| si:ch1073-110a20.3 | 0.57 |
| si:dkey-22o12.5 | 0.51 |
| atp10d | 0.50 |
Gene Ontology
Download
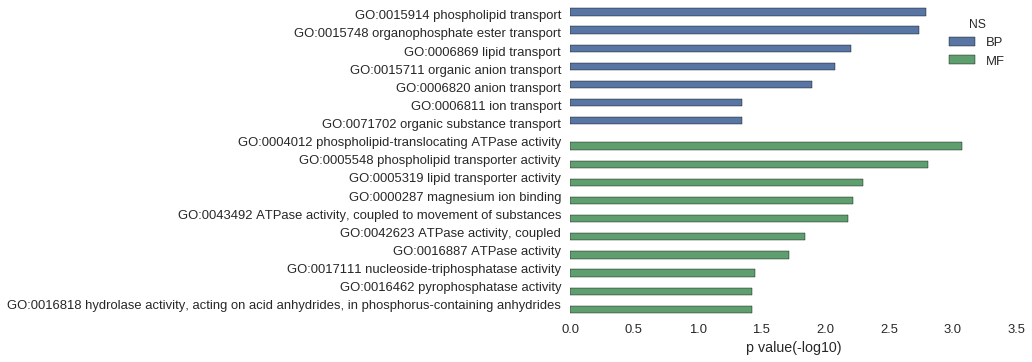
| GO | P value |
|---|---|
| GO:0015914 | 1.63e-03 |
| GO:0015748 | 1.85e-03 |
| GO:0006869 | 6.32e-03 |
| GO:0015711 | 8.39e-03 |
| GO:0006820 | 1.27e-02 |
| GO:0006811 | 4.49e-02 |
| GO:0071702 | 4.49e-02 |
| GO:0004012 | 8.53e-04 |
| GO:0005548 | 1.56e-03 |
| GO:0005319 | 5.05e-03 |
| GO:0000287 | 6.11e-03 |
| GO:0043492 | 6.68e-03 |
| GO:0042623 | 1.44e-02 |
| GO:0016887 | 1.93e-02 |
| GO:0017111 | 3.55e-02 |
| GO:0016462 | 3.72e-02 |
| GO:0016818 | 3.74e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT01065 | NONHSAT098858; NONHSAT216814; NONHSAT216815; NONHSAT199265; | Collinearity with conserved coding gene |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG00717
ACAAGGAGGGCAAGACACAAAAGATCATTGCAAAAGAGGCCGGCGGTTCACGAAGATCTGTGTCCAAGCA
CATTAATTGTGGTAGGACAAAGTGTACAAGCAATAGGCATGACCGCGCCCTGGAGAGATCTGTAAAACAA
AACGCATTTAAAAATGTGGAGGAAATTCACAAAGAGTGGACTGCAGCTGGAGTCAGGAAGAGAGGAGAGG
CACACAATCCACGTTGCTTGATGTCCAGTGTAAAGTTTCTGCGGTCAGTGATGGTTTGGGCTGCCATGTC
ATCTGCTGGTGTTGCTCCACTGTGTTTTCTGAGGTCCAAGGTCAACACAGCTCTATACCAGGACATTTTA
GAGCACTACATGCTTCCCATTGCTGACCAACTTTATGGAGATGCGGATTTAATTTTCCAACAGGACTTGG
CACCTGCACACAGTACCAAAGCTACCAGTACCTGCTTTAAGGACCGTGGTATCTCTGTTCTTAATTTGCC
AGTAAACTCGCCTAACCTTAACCCCACAGGAAATCTATGGGCTATCGTGAAGAGGAAGATGCGGAATGCC
AGACCCAACAATGCAGAGAAGCTGAAGGCCACTATCAGAGCCACCTGGGCTCTCAGAAAACCTGAGCAGT
GCTGAACACAGACTGATCCACTCCATGACAAGCCGCATTGCTGCAGCAATTCAGACAAAAGGAGCCCACA
CTGACTATTGA
ACAAGGAGGGCAAGACACAAAAGATCATTGCAAAAGAGGCCGGCGGTTCACGAAGATCTGTGTCCAAGCA
CATTAATTGTGGTAGGACAAAGTGTACAAGCAATAGGCATGACCGCGCCCTGGAGAGATCTGTAAAACAA
AACGCATTTAAAAATGTGGAGGAAATTCACAAAGAGTGGACTGCAGCTGGAGTCAGGAAGAGAGGAGAGG
CACACAATCCACGTTGCTTGATGTCCAGTGTAAAGTTTCTGCGGTCAGTGATGGTTTGGGCTGCCATGTC
ATCTGCTGGTGTTGCTCCACTGTGTTTTCTGAGGTCCAAGGTCAACACAGCTCTATACCAGGACATTTTA
GAGCACTACATGCTTCCCATTGCTGACCAACTTTATGGAGATGCGGATTTAATTTTCCAACAGGACTTGG
CACCTGCACACAGTACCAAAGCTACCAGTACCTGCTTTAAGGACCGTGGTATCTCTGTTCTTAATTTGCC
AGTAAACTCGCCTAACCTTAACCCCACAGGAAATCTATGGGCTATCGTGAAGAGGAAGATGCGGAATGCC
AGACCCAACAATGCAGAGAAGCTGAAGGCCACTATCAGAGCCACCTGGGCTCTCAGAAAACCTGAGCAGT
GCTGAACACAGACTGATCCACTCCATGACAAGCCGCATTGCTGCAGCAATTCAGACAAAAGGAGCCCACA
CTGACTATTGA
