ZFLNCG04538
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 0.82 |
| SRR1299127 | caudal fin | Two days time after treatment | 0.76 |
| SRR700537 | heart | Gata4 morpholino | 0.71 |
| SRR516133 | skin | male and 5 month | 0.62 |
| SRR658539 | bud | Gata5/6 morphant | 0.56 |
| SRR594769 | blood | normal | 0.55 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 0.54 |
| SRR658537 | bud | Gata6 morphant | 0.49 |
| SRR658547 | 6 somite | Gata5/6 morphant | 0.46 |
| SRR1021213 | sphere stage | normal | 0.44 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| cep57 | 0.52 |
| LOC100537540 | 0.51 |
| kri1 | 0.50 |
Gene Ontology
Download
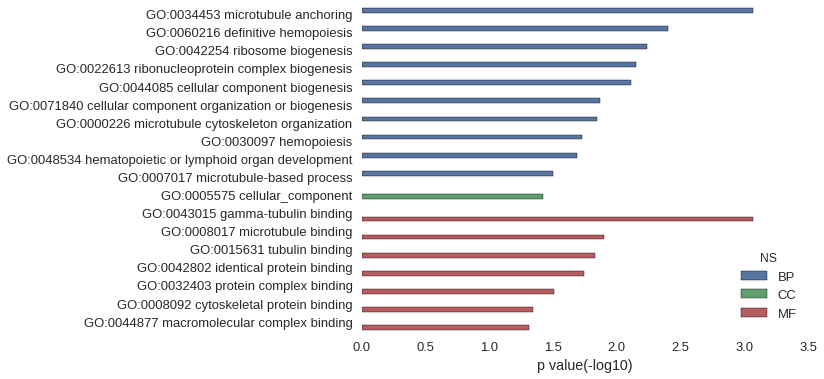
| GO | P value |
|---|---|
| GO:0034453 | 8.53e-04 |
| GO:0060216 | 3.98e-03 |
| GO:0042254 | 5.82e-03 |
| GO:0022613 | 7.09e-03 |
| GO:0044085 | 7.80e-03 |
| GO:0071840 | 1.35e-02 |
| GO:0000226 | 1.43e-02 |
| GO:0030097 | 1.88e-02 |
| GO:0048534 | 2.06e-02 |
| GO:0007017 | 3.17e-02 |
| GO:0005575 | 3.80e-02 |
| GO:0043015 | 8.53e-04 |
| GO:0008017 | 1.25e-02 |
| GO:0015631 | 1.47e-02 |
| GO:0042802 | 1.80e-02 |
| GO:0032403 | 3.10e-02 |
| GO:0008092 | 4.55e-02 |
| GO:0044877 | 4.86e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04538
AAAAAGAAAGTCAAAATAAACCAATAGGACACAACAAGGAAACACTATTATTGAAATTTTGTTTATTTAA
TGACATTGGGAAATGGAAATTGCTGAGACTTTCACTTCAGTGTGTGGATGTACTTCTAACTGAAACTAAA
GGGGCGGGACTTCTTTTTGCACATCATTCCCTCATAGCAAACTAATAGTAAGAGGGCCATGGTTGCAAAT
ATTGTGGCTTAAGCAGTCAAACTGACATCTTCAGAGAATGGCCACAACTCCAAACCCAAAGACTTTGACT
TGACTATAGAAACTTTATTTTTTTAATGGATTAATTGCCTCAAATGAACGGTTCACTTAAAGAATAACAA
TGTACTCTAGCCAAAAAAAAAAACACTGTAAAGTTCACAGACTTTTAGCTGTAAATTAGCAAATTGACAT
GACATAAATCAAAATAAAGGCTCAGTAAATGTTGAGAGCACAACTTGGCATCAGTTTGTGTGCGAACTAA
CCAATTGTTTTGTCCGCTATTACTCCAACCGCCAGGCGGAAATTACAAATCGCCGTTTCATTCCAGCGAC
GAATCATCTATATGAACGAAGGAAAACAGGAAAACAAGACAGAAGATGAGACGGAGAGGCAGAGGGGGAG
AAAAAAAATCAGTCGAAAATCAGAAAAGATGAATCTGTCTTTCTCGTCTAATGAGCGTCTGATTGAATAC
TTCAGAGGGAGAACATGTAGTCATTAAGTTCAGCTCCTCAAAGACGACCACCAAAAAAATCTAATTACAA
TAAAACAGACATAACATAGCTGTGAAGCAGCCACTTACGGCTAATCATATCAATTACTGCGCCCGGGGCT
CCTCAGAGACACAGAGTGATTGTTTGAAGAGAAAAATTGTATTTGGTGAGCCCGAGACACTCACTCACAG
CAAACCATCCATTACTCTCCATTCACTTTCCTCTGCA
AAAAAGAAAGTCAAAATAAACCAATAGGACACAACAAGGAAACACTATTATTGAAATTTTGTTTATTTAA
TGACATTGGGAAATGGAAATTGCTGAGACTTTCACTTCAGTGTGTGGATGTACTTCTAACTGAAACTAAA
GGGGCGGGACTTCTTTTTGCACATCATTCCCTCATAGCAAACTAATAGTAAGAGGGCCATGGTTGCAAAT
ATTGTGGCTTAAGCAGTCAAACTGACATCTTCAGAGAATGGCCACAACTCCAAACCCAAAGACTTTGACT
TGACTATAGAAACTTTATTTTTTTAATGGATTAATTGCCTCAAATGAACGGTTCACTTAAAGAATAACAA
TGTACTCTAGCCAAAAAAAAAAACACTGTAAAGTTCACAGACTTTTAGCTGTAAATTAGCAAATTGACAT
GACATAAATCAAAATAAAGGCTCAGTAAATGTTGAGAGCACAACTTGGCATCAGTTTGTGTGCGAACTAA
CCAATTGTTTTGTCCGCTATTACTCCAACCGCCAGGCGGAAATTACAAATCGCCGTTTCATTCCAGCGAC
GAATCATCTATATGAACGAAGGAAAACAGGAAAACAAGACAGAAGATGAGACGGAGAGGCAGAGGGGGAG
AAAAAAAATCAGTCGAAAATCAGAAAAGATGAATCTGTCTTTCTCGTCTAATGAGCGTCTGATTGAATAC
TTCAGAGGGAGAACATGTAGTCATTAAGTTCAGCTCCTCAAAGACGACCACCAAAAAAATCTAATTACAA
TAAAACAGACATAACATAGCTGTGAAGCAGCCACTTACGGCTAATCATATCAATTACTGCGCCCGGGGCT
CCTCAGAGACACAGAGTGATTGTTTGAAGAGAAAAATTGTATTTGGTGAGCCCGAGACACTCACTCACAG
CAAACCATCCATTACTCTCCATTCACTTTCCTCTGCA
