ZFLNCG06401
Basic Information
Chromesome: chr10
Start: 29136281
End: 29136952
Transcript: ZFLNCT09813
Known as: ENSDARG00000094958
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035239 | liver | transgenic mCherry control | 22.73 |
| SRR700534 | heart | control morpholino | 22.53 |
| SRR700537 | heart | Gata4 morpholino | 8.93 |
| SRR658539 | bud | Gata5/6 morphant | 7.07 |
| SRR658535 | bud | Gata5 morphant | 6.93 |
| SRR658543 | 6 somite | Gata5 morphan | 6.13 |
| SRR1562530 | liver | normal | 6.10 |
| SRR941753 | posterior pectoral fin | normal | 4.53 |
| SRR658537 | bud | Gata6 morphant | 4.15 |
| SRR658545 | 6 somite | Gata6 morphant | 4.09 |
Express in tissues
Correlated coding gene
Download
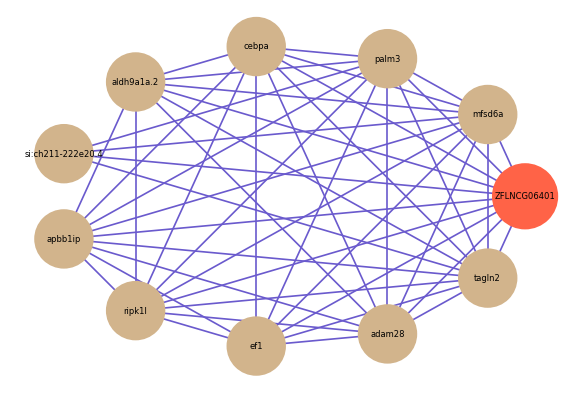
| gene | correlation coefficent |
|---|---|
| mfsd6a | 0.62 |
| palm3 | 0.62 |
| cebpa | 0.61 |
| aldh9a1a.2 | 0.60 |
| si:ch211-222e20.4 | 0.60 |
| apbb1ip | 0.60 |
| ripk1l | 0.60 |
| ef1 | 0.60 |
| adam28 | 0.58 |
| tagln2 | 0.58 |
Gene Ontology
Download
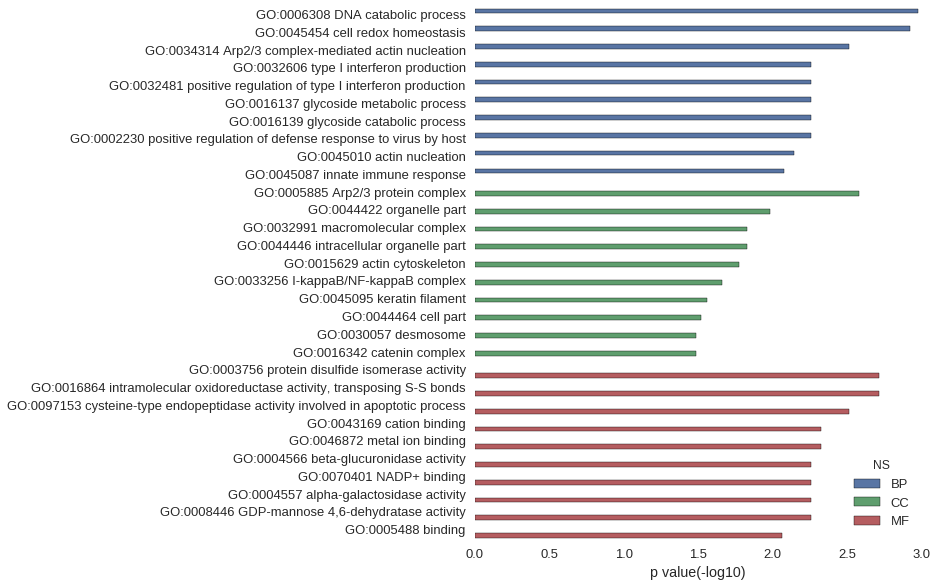
| GO | P value |
|---|---|
| GO:0006308 | 1.06e-03 |
| GO:0045454 | 1.20e-03 |
| GO:0034314 | 3.04e-03 |
| GO:0032606 | 5.54e-03 |
| GO:0032481 | 5.54e-03 |
| GO:0016137 | 5.54e-03 |
| GO:0016139 | 5.54e-03 |
| GO:0002230 | 5.54e-03 |
| GO:0045010 | 7.12e-03 |
| GO:0045087 | 8.31e-03 |
| GO:0005885 | 2.64e-03 |
| GO:0044422 | 1.04e-02 |
| GO:0032991 | 1.48e-02 |
| GO:0044446 | 1.48e-02 |
| GO:0015629 | 1.69e-02 |
| GO:0033256 | 2.20e-02 |
| GO:0045095 | 2.74e-02 |
| GO:0044464 | 3.03e-02 |
| GO:0030057 | 3.28e-02 |
| GO:0016342 | 3.28e-02 |
| GO:0003756 | 1.93e-03 |
| GO:0016864 | 1.93e-03 |
| GO:0097153 | 3.04e-03 |
| GO:0043169 | 4.70e-03 |
| GO:0046872 | 4.72e-03 |
| GO:0004566 | 5.54e-03 |
| GO:0070401 | 5.54e-03 |
| GO:0004557 | 5.54e-03 |
| GO:0008446 | 5.54e-03 |
| GO:0005488 | 8.62e-03 |
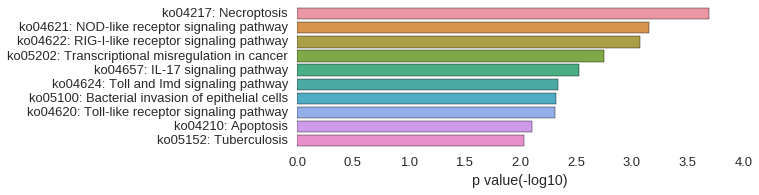
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06401
TCTGTTAGCATTTTCTGATCTGCTGAATTTGATGCTTTTTTGGGTTTGGGTTCCTGAGTAGTTGACGTGA
CAGGTGATTGTGTCTCCTCCACTCCAGTCAGGTTTGCACAGCTGCAGTTCACTATTATCAGCATTAAATG
TGGACACAAGTGGTAACATCTGACCTCCTCGGCTCCAGGTGAAGTTGGCCAGATCAGAGTCAAGAGAGCA
GAGCAGATGAACACAGCTGTCATTAGTGCTGTTCAGTGGCTTCAGGGACAGACCTAGACATTAGATTCAT
CAAGGTAAGATGATATTCAACATACAGACAATTCAGAATCACTTATGAACAGCAAAATCACCTGCGACTT
GGAGAATCAGAGATGAGCTTTCTTCCACTGACGGAGGTGGAATTAATGTAGTTGTTTTACAGCTGTATTT
ACCAGAGTCACCAGGCTGAACATTTTTGATCTCTAATGTAACAGGCTTACTGGTTATTGTAAAATGAGGA
AAGCAATCTTTCAAGGGAGATACAAGGGAGTTGGACATATCAATGCTCCATGTACAAATAAAATTGCTAC
TGGTTGTATTGATGCTCCACTCTACTGTAACCTGTAGACTCTCTACAGAAGGTAAATGGCAGGGAAGATG
GACAGTGGCTCCTTCCACTGCTCTGATAACTTTCTTTGCAT
TCTGTTAGCATTTTCTGATCTGCTGAATTTGATGCTTTTTTGGGTTTGGGTTCCTGAGTAGTTGACGTGA
CAGGTGATTGTGTCTCCTCCACTCCAGTCAGGTTTGCACAGCTGCAGTTCACTATTATCAGCATTAAATG
TGGACACAAGTGGTAACATCTGACCTCCTCGGCTCCAGGTGAAGTTGGCCAGATCAGAGTCAAGAGAGCA
GAGCAGATGAACACAGCTGTCATTAGTGCTGTTCAGTGGCTTCAGGGACAGACCTAGACATTAGATTCAT
CAAGGTAAGATGATATTCAACATACAGACAATTCAGAATCACTTATGAACAGCAAAATCACCTGCGACTT
GGAGAATCAGAGATGAGCTTTCTTCCACTGACGGAGGTGGAATTAATGTAGTTGTTTTACAGCTGTATTT
ACCAGAGTCACCAGGCTGAACATTTTTGATCTCTAATGTAACAGGCTTACTGGTTATTGTAAAATGAGGA
AAGCAATCTTTCAAGGGAGATACAAGGGAGTTGGACATATCAATGCTCCATGTACAAATAAAATTGCTAC
TGGTTGTATTGATGCTCCACTCTACTGTAACCTGTAGACTCTCTACAGAAGGTAAATGGCAGGGAAGATG
GACAGTGGCTCCTTCCACTGCTCTGATAACTTTCTTTGCAT