ZFLNCG06505
Basic Information
Chromesome: chr10
Start: 41794800
End: 41795803
Transcript: ZFLNCT09972
Known as: ENSDARG00000092993
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 3.31 |
| ERR023146 | head kidney | normal | 0.60 |
| SRR1562533 | testis | normal | 0.27 |
| SRR1647679 | head kidney | normal | 0.23 |
| SRR1647680 | head kidney | SVCV treatment | 0.22 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 0.20 |
| SRR1205166 | 5dpf | transgenic sqET20 and neomycin treated 3h | 0.17 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 0.16 |
| SRR630463 | 5 dpf | normal | 0.16 |
| SRR942772 | embryo | myd88+/+ and infection with Mycobacterium marinum | 0.15 |
Express in tissues
Gene Ontology
Download
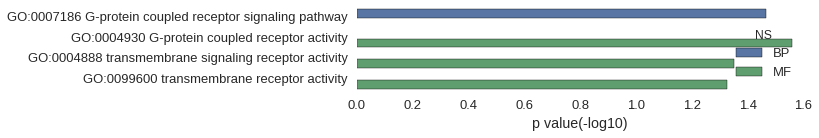
| GO | P value |
|---|---|
| GO:0007186 | 3.41e-02 |
| GO:0004930 | 2.76e-02 |
| GO:0004888 | 4.45e-02 |
| GO:0099600 | 4.70e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT09972 | ENST00000486251; NONHSAT125668; NONHSAT104458; NONHSAT024249; NONHSAT125667; | ENSMUST00000172211; ENSMUST00000174168; ENSMUST00000173181; ENSMUST00000172477; NONMMUT029605; NONMMUT029606; NONMMUT029607; | Collinearity with conserved coding gene |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06505
TAATGTTCTGTAAAAATATTGATCAGAGAAGATGCAGAATGAAAAATTTTCAGGGTTAAAATGAGTTTAG
CTGTCTTCTTAAACCAGGGGTAAAATAAAGCATAAATCAGAGGATTCAGACCTGAGTTGGCACATACAGC
AAATAATAAACCATTTATAGTTGTGGAGGAGTTTACAGTTATAGAGCAGATGAAATATGGAATATAGCAA
AGCAGATAAACTAACACAATGATTCCTAGAGTCAGAGCAGCTTTACTCTCAGATTTCCTCTTCACTAAAC
CCTCCGTTACACATTTACCACCCTTCATCAGAGAGTTTATAACCTTCACTTGCTGATGAACGACATAGAA
GATCCTTAAATATAAAGAGATGATCAGCAAACAAGGAAAAATAAAAGACAAGAACAGATCAGTCACTACC
CAATTGAAACTTGACATGAACAAACACTCCCCATAACACACGTCTGTTCTGTGTGAAGTGCCAAAATATC
CATTATTGATTAAAAGGGCAGTGTTATAAGCTGATGAACAAACCCAGCTCAGACAGATACTCATAAGTGT
TTTAGTGATGGTTATTTTCTGTGGGTACAGTAAAGGGTGACACACAGCCACATAACGATCAACAGCAATT
AAAACCAAATTACTAAGAGATGCTGCAATAAGTAATGACACAAATAATAAATGAAGTCCACAGAAAGTGT
CTCCAAAATACCAACACGTCTCAATCATTCTGATGGCCTCTACAGGCATGACGAGTCCAACAAGCAGATC
AGTTACAGCCAGAGAGAGAATAATCATGTTGGTTGGAGTGTGAAGCTTCTTGAAGTGAGAGATGGAGATG
ATCACCAGCAGGTTCAGAAACACAGTCCATGCTGACAGCAATGACACAAACAAGTATATGATAACAAATC
TATGACTGGAGCGTCTCTCCTTGACACATGATGAGTTGATGTCAGGAAAGCAGTATTGAGTCTCCAGATC
CTCTGTCTCATAGGCCATGAGTG
TAATGTTCTGTAAAAATATTGATCAGAGAAGATGCAGAATGAAAAATTTTCAGGGTTAAAATGAGTTTAG
CTGTCTTCTTAAACCAGGGGTAAAATAAAGCATAAATCAGAGGATTCAGACCTGAGTTGGCACATACAGC
AAATAATAAACCATTTATAGTTGTGGAGGAGTTTACAGTTATAGAGCAGATGAAATATGGAATATAGCAA
AGCAGATAAACTAACACAATGATTCCTAGAGTCAGAGCAGCTTTACTCTCAGATTTCCTCTTCACTAAAC
CCTCCGTTACACATTTACCACCCTTCATCAGAGAGTTTATAACCTTCACTTGCTGATGAACGACATAGAA
GATCCTTAAATATAAAGAGATGATCAGCAAACAAGGAAAAATAAAAGACAAGAACAGATCAGTCACTACC
CAATTGAAACTTGACATGAACAAACACTCCCCATAACACACGTCTGTTCTGTGTGAAGTGCCAAAATATC
CATTATTGATTAAAAGGGCAGTGTTATAAGCTGATGAACAAACCCAGCTCAGACAGATACTCATAAGTGT
TTTAGTGATGGTTATTTTCTGTGGGTACAGTAAAGGGTGACACACAGCCACATAACGATCAACAGCAATT
AAAACCAAATTACTAAGAGATGCTGCAATAAGTAATGACACAAATAATAAATGAAGTCCACAGAAAGTGT
CTCCAAAATACCAACACGTCTCAATCATTCTGATGGCCTCTACAGGCATGACGAGTCCAACAAGCAGATC
AGTTACAGCCAGAGAGAGAATAATCATGTTGGTTGGAGTGTGAAGCTTCTTGAAGTGAGAGATGGAGATG
ATCACCAGCAGGTTCAGAAACACAGTCCATGCTGACAGCAATGACACAAACAAGTATATGATAACAAATC
TATGACTGGAGCGTCTCTCCTTGACACATGATGAGTTGATGTCAGGAAAGCAGTATTGAGTCTCCAGATC
CTCTGTCTCATAGGCCATGAGTG