ZFLNCG08597
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 6.82 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 3.71 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 3.53 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 3.24 |
| SRR942767 | embryo | myd88-/- and infection with Mycobacterium marinum | 0.91 |
| SRR1342219 | embryo | marco morpholino and infection with Mycobacterium marinum | 0.64 |
| SRR1205154 | 5dpf | transgenic sqET20 | 0.61 |
| SRR1565809 | brain | normal | 0.59 |
| SRR776508 | embryo | miR-146 knockdown | 0.57 |
| SRR891510 | muscle | normal | 0.48 |
Express in tissues
Correlated coding gene
Download
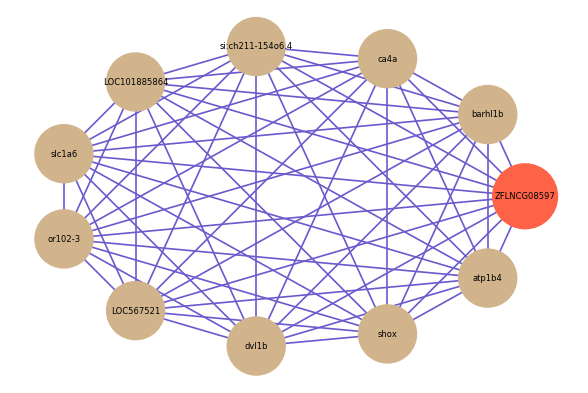
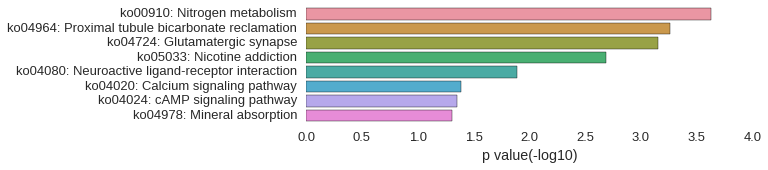
| gene | correlation coefficent |
|---|---|
| barhl1b | 0.54 |
| ca4a | 0.54 |
| si:ch211-154o6.4 | 0.53 |
| LOC101885864 | 0.52 |
| slc1a6 | 0.52 |
| or102-3 | 0.52 |
| LOC567521 | 0.52 |
| dvl1b | 0.52 |
| shox | 0.52 |
| atp1b4 | 0.52 |
Gene Ontology
Download
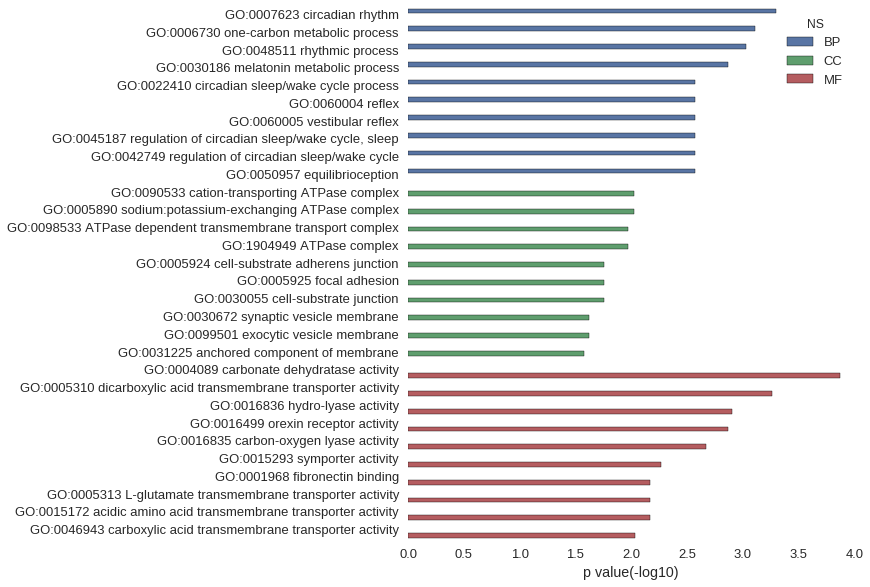
| GO | P value |
|---|---|
| GO:0007623 | 5.09e-04 |
| GO:0006730 | 7.85e-04 |
| GO:0048511 | 9.44e-04 |
| GO:0030186 | 1.35e-03 |
| GO:0022410 | 2.70e-03 |
| GO:0060004 | 2.70e-03 |
| GO:0060005 | 2.70e-03 |
| GO:0045187 | 2.70e-03 |
| GO:0042749 | 2.70e-03 |
| GO:0050957 | 2.70e-03 |
| GO:0090533 | 9.42e-03 |
| GO:0005890 | 9.42e-03 |
| GO:0098533 | 1.08e-02 |
| GO:1904949 | 1.08e-02 |
| GO:0005924 | 1.74e-02 |
| GO:0005925 | 1.74e-02 |
| GO:0030055 | 1.74e-02 |
| GO:0030672 | 2.40e-02 |
| GO:0099501 | 2.40e-02 |
| GO:0031225 | 2.67e-02 |
| GO:0004089 | 1.34e-04 |
| GO:0005310 | 5.51e-04 |
| GO:0016836 | 1.24e-03 |
| GO:0016499 | 1.35e-03 |
| GO:0016835 | 2.12e-03 |
| GO:0015293 | 5.38e-03 |
| GO:0001968 | 6.73e-03 |
| GO:0005313 | 6.73e-03 |
| GO:0015172 | 6.73e-03 |
| GO:0046943 | 9.33e-03 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08597
ACACACACACACACACACACACACACACCAAACACACACACACACACACACACAAAAACACACAAAGGAT
TTGTCACATATAAACATCATGTGGTCACGATGATTTACTGCTTCAAAACAAAAGAGATTAAAGATGCATT
AAAAAGGAAAATGAGACACAAGAGAGTCAGCATGAAAGTTAAAATGATGTAATGCTTTACTATAATATAG
TGTCATTCATGCATGCCTCGTTTTTTTCTATTTCTGCAACAAAGGTTGTACATTTTTGCAATTAATAGAC
ATAAATGCAAAAATAAATGTTTAAAAGTATGCTATACTTTAATAATGAAAAATAGAAATGGGCTTTTTAT
CTCACATTTTCTTTTTCACTTGTATTATTAGAGTGCAGGCTACAGTGAAGTACTACTAAAATGTTTTATA
CTTTGAACCTTACTTTGAAATTTTTAGCTTGTTTTTAGATTGTTATGATCAGATGAATGGCATTTCATTT
AGTTCATTTATTTCATTGTTTTTATAATTGTGTGTTATTGAAGTGTGTAGGGATATTCTTCTGAGATAGC
TTTATAATGTTTGTTGTTTTAGTTG
ACACACACACACACACACACACACACACCAAACACACACACACACACACACACAAAAACACACAAAGGAT
TTGTCACATATAAACATCATGTGGTCACGATGATTTACTGCTTCAAAACAAAAGAGATTAAAGATGCATT
AAAAAGGAAAATGAGACACAAGAGAGTCAGCATGAAAGTTAAAATGATGTAATGCTTTACTATAATATAG
TGTCATTCATGCATGCCTCGTTTTTTTCTATTTCTGCAACAAAGGTTGTACATTTTTGCAATTAATAGAC
ATAAATGCAAAAATAAATGTTTAAAAGTATGCTATACTTTAATAATGAAAAATAGAAATGGGCTTTTTAT
CTCACATTTTCTTTTTCACTTGTATTATTAGAGTGCAGGCTACAGTGAAGTACTACTAAAATGTTTTATA
CTTTGAACCTTACTTTGAAATTTTTAGCTTGTTTTTAGATTGTTATGATCAGATGAATGGCATTTCATTT
AGTTCATTTATTTCATTGTTTTTATAATTGTGTGTTATTGAAGTGTGTAGGGATATTCTTCTGAGATAGC
TTTATAATGTTTGTTGTTTTAGTTG