ZFLNCG08599
Basic Information
Chromesome: chr15
Start: 30777209
End: 30778118
Transcript: ZFLNCT13247
Known as: ENSDARG00000092217
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 2.97 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 2.72 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 2.63 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 2.15 |
| ERR023144 | brain | normal | 1.15 |
| SRR726540 | 5 dpf | normal | 0.44 |
| SRR1188148 | embryo | Control PBS 0.5 hpi | 0.37 |
| SRR1565809 | brain | normal | 0.36 |
| SRR942769 | embryo | myd88+/+ | 0.33 |
| SRR1565811 | brain | normal | 0.29 |
Express in tissues
Correlated coding gene
Download
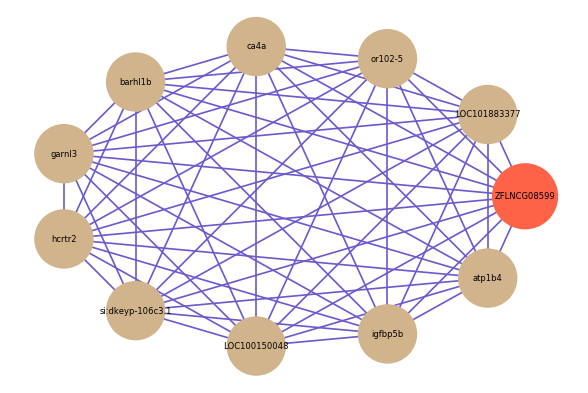
| gene | correlation coefficent |
|---|---|
| LOC101883377 | 0.61 |
| or102-5 | 0.61 |
| ca4a | 0.60 |
| barhl1b | 0.60 |
| garnl3 | 0.60 |
| hcrtr2 | 0.60 |
| si:dkeyp-106c3.1 | 0.59 |
| LOC100150048 | 0.59 |
| igfbp5b | 0.58 |
| atp1b4 | 0.58 |
Gene Ontology
Download
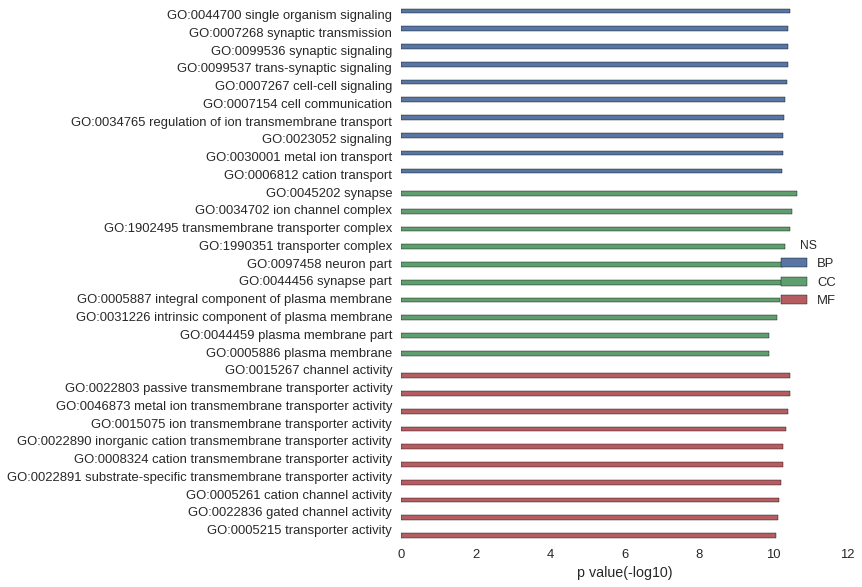
| GO | P value |
|---|---|
| GO:0044700 | 3.57e-11 |
| GO:0007268 | 4.07e-11 |
| GO:0099536 | 4.07e-11 |
| GO:0099537 | 4.07e-11 |
| GO:0007267 | 4.20e-11 |
| GO:0007154 | 4.99e-11 |
| GO:0034765 | 5.05e-11 |
| GO:0023052 | 5.33e-11 |
| GO:0030001 | 5.65e-11 |
| GO:0006812 | 5.78e-11 |
| GO:0045202 | 2.28e-11 |
| GO:0034702 | 3.17e-11 |
| GO:1902495 | 3.51e-11 |
| GO:1990351 | 4.75e-11 |
| GO:0097458 | 5.56e-11 |
| GO:0044456 | 5.99e-11 |
| GO:0005887 | 6.48e-11 |
| GO:0031226 | 8.07e-11 |
| GO:0044459 | 1.30e-10 |
| GO:0005886 | 1.33e-10 |
| GO:0015267 | 3.54e-11 |
| GO:0022803 | 3.54e-11 |
| GO:0046873 | 3.93e-11 |
| GO:0015075 | 4.50e-11 |
| GO:0022890 | 5.35e-11 |
| GO:0008324 | 5.59e-11 |
| GO:0022891 | 6.05e-11 |
| GO:0005261 | 7.03e-11 |
| GO:0022836 | 7.66e-11 |
| GO:0005215 | 8.43e-11 |
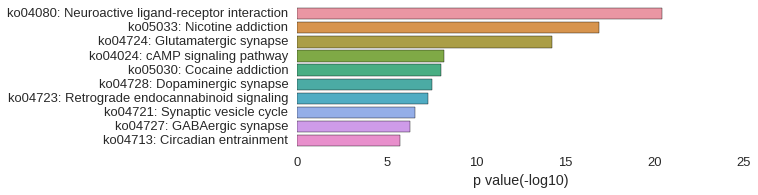
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08599
CTACCAAAACAGAAACAGCACAGCAATAATCACTGAGTTTGATCTGCGAGGTTTTCCAGGCCTCCTTCGT
GAGTACAGACTTGTCACAGCTTTATTTTCATCAACTGGGAATATTTTCATCATTGTCTTTGTTATGCATT
AACAAACCCAACCAGGATAATCTTTTGTAACCTCGTCGTGGCAGATCTTGCACTTGGAACAACAACAACC
TACCCAGGAGTCTAAATACTAAATACTGAATGTCTGACAAAATCAATTTATTCTGTGGCTTCTTCAGGCA
GATGGTTTTGTCCATTTTTTGGGGCTATAAATTCATTTTTCATGGCACTGATGGCTTTTGATCGATTTAC
AGCTACATTTAATCCCCTTAGATATTATACCCTGATCAAAAACTTCACTATTCATTTCATGTGTGTGTCT
GTTTGGACAGCAAACATGCTGCATATAGTTGGAGTCACTCTCTTATCCCTCAGTGTGAAATTTTGTGGGT
CAAATGTCATTCCTCACTGCTACTGTGATCATGTCTCTATAAGCAAACTGGCATGTGGAGATGCTGCTAC
CATGAAAGTCACAAGTACAGCCATTGCCATGTTTGGTCTGTGGGGTCCTTTACGTTTCATCTTATTCTCG
TATGTCTCTACTATCGTCTCTGTATCAAAAATCTCCAGGCCTACAGGTCGACATAAACCATTCTCCACAT
GTACACCACAGTTACTGATCATATGTTTGTATTATCTACCCAGAACATTTGTGTATTTAACACACATTGC
TGGATATGAATTTAGTAATGACATTTGCATCGAAGTGTCCATGATGTACAGTCTCTTCCCTGCTGTTATC
AGCCGTCTTATATATTGCTTCAGGACTAAAGTGATTAAAGAGGCGATATTTAAAAGAGTTACTAAAACA
CTACCAAAACAGAAACAGCACAGCAATAATCACTGAGTTTGATCTGCGAGGTTTTCCAGGCCTCCTTCGT
GAGTACAGACTTGTCACAGCTTTATTTTCATCAACTGGGAATATTTTCATCATTGTCTTTGTTATGCATT
AACAAACCCAACCAGGATAATCTTTTGTAACCTCGTCGTGGCAGATCTTGCACTTGGAACAACAACAACC
TACCCAGGAGTCTAAATACTAAATACTGAATGTCTGACAAAATCAATTTATTCTGTGGCTTCTTCAGGCA
GATGGTTTTGTCCATTTTTTGGGGCTATAAATTCATTTTTCATGGCACTGATGGCTTTTGATCGATTTAC
AGCTACATTTAATCCCCTTAGATATTATACCCTGATCAAAAACTTCACTATTCATTTCATGTGTGTGTCT
GTTTGGACAGCAAACATGCTGCATATAGTTGGAGTCACTCTCTTATCCCTCAGTGTGAAATTTTGTGGGT
CAAATGTCATTCCTCACTGCTACTGTGATCATGTCTCTATAAGCAAACTGGCATGTGGAGATGCTGCTAC
CATGAAAGTCACAAGTACAGCCATTGCCATGTTTGGTCTGTGGGGTCCTTTACGTTTCATCTTATTCTCG
TATGTCTCTACTATCGTCTCTGTATCAAAAATCTCCAGGCCTACAGGTCGACATAAACCATTCTCCACAT
GTACACCACAGTTACTGATCATATGTTTGTATTATCTACCCAGAACATTTGTGTATTTAACACACATTGC
TGGATATGAATTTAGTAATGACATTTGCATCGAAGTGTCCATGATGTACAGTCTCTTCCCTGCTGTTATC
AGCCGTCTTATATATTGCTTCAGGACTAAAGTGATTAAAGAGGCGATATTTAAAAGAGTTACTAAAACA