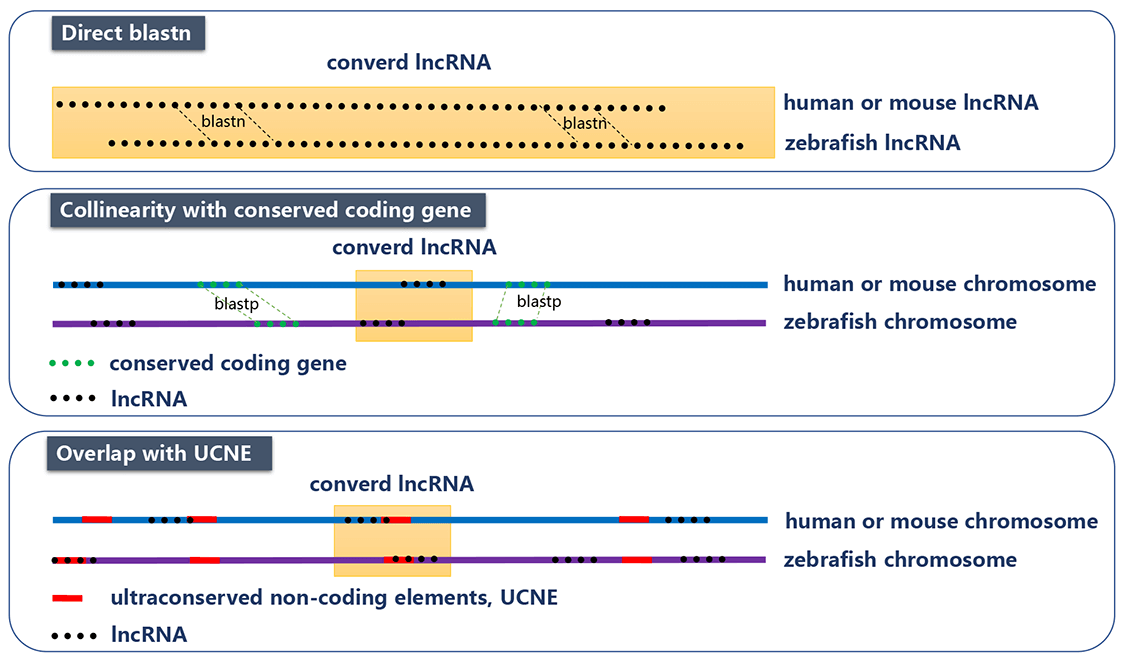
We used three methods (that are direct BLASTN, collinearity with conserved coding gene, and overlap with multi-species ultra-conserved non-coding elements (UCNE)) to find the counterparts of zebrafish lncRNAs in human or mouse. In direct comparison of zebrafish lncRNA and human/mouse lncRNA with BLASTN, bidirectional best hits using a relatively non-stringent threshold (E-value<=10-5) were considered as orthologs. In collinearity method, we compared the coding genes of zebrafish with those of human or mouse using BLASTP as anchor points. We assumed that those lncRNAs with more than 5 anchor points in the 20k upstream/downstream region are orthologs. In UCNE method, if two lncRNAs from different species overlap with at least one UCNE, as another anchor point, they were considered as orthologs. Finally, we obtained 2,156 zebrafish lncRNA genes that have the counterpart in human or mouse.