GCEN: an easy-to-use toolkit for Gene Co-Expression Network analysis and lncRNAs annotation
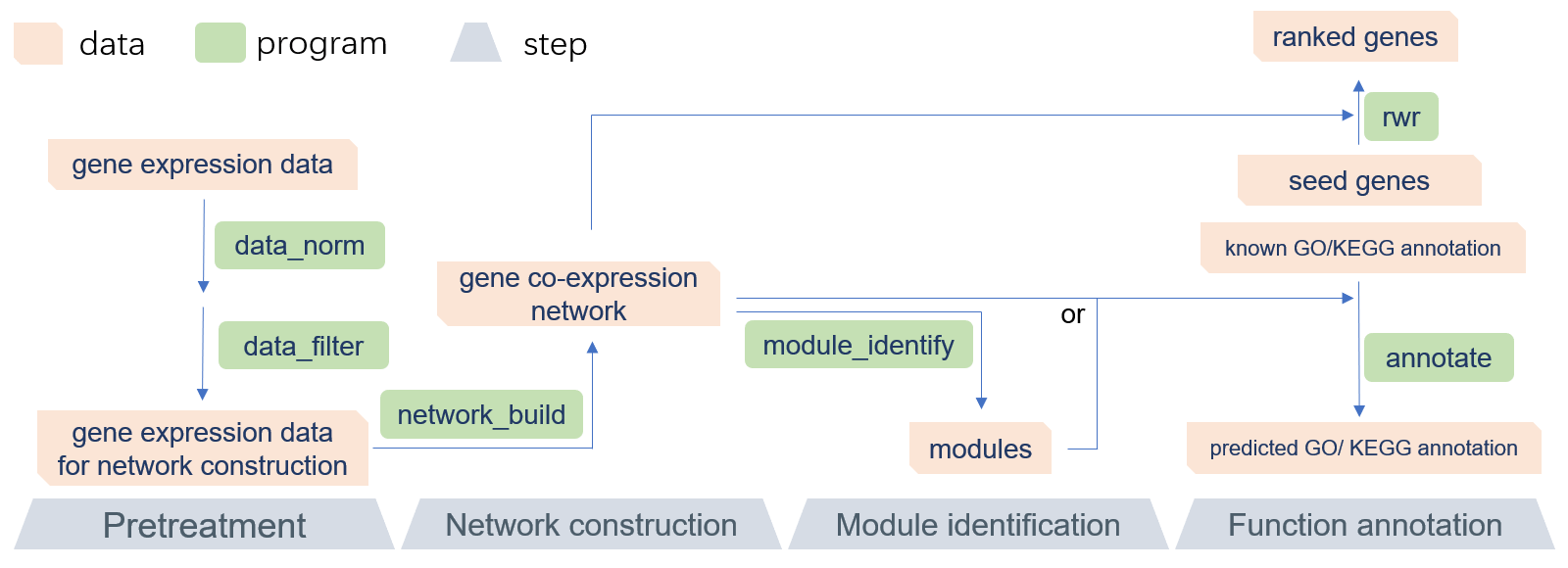
GCEN workflow
GCEN is a command-line toolkit that allows biologists to easily build gene co-expression network and predict gene function, especially in RNA-Seq research or lncRNAs annotation. GCEN is primarily designed to be used in lncRNAs annotation, but is not limited to those scenarios. It is an efficient and easy-to-use solution that will allow everyone to perform gene co-expression network analysis without sophisticated programming skills. The recommended pipeline consists of four parts: data pretreatment, network construction, module identification, and function annotation. A README file and sample data are included in the software package. Because of its modular design, the GCEN can be easily integrated into another pipeline. Also, the multithreaded implementation of GCEN makes it fast and efficient for RNA-Seq data.
Download
GCEN is an open source software under the GPLv3 license. We provide source code and pre-built binaries. GCEN only supports 64-bit operating system and has been tested in Ubuntu 18.04 (Bionic Beaver), Ubuntu 20.04 (Focal Fossa), Windows 7, Windows 10, macOS 10.15 (Catalina) and macOS 11.0 (Big Sur).
gcen-0.6.3-linux-x86_64.tar.gz
gcen-0.6.3-macOS-x86_64.tar.gz
For Linux and macOS user, you can install GCEN with conda. Since bioconda does not support Windows, Windows users can download the pre-built binaries directly from our website.
conda install gcen -c biocondaContact
Wen Chen, Ph.D. Candidate
Email: chenwen@biochen.org
Citation
Wen Chen, Jing Li, Shulan Huang, Xiaodeng Li, Xuan Zhan, Xiang Hu, Shuanglin Xiang, Changning Liu. GCEN: An Easy-to-Use Toolkit for Gene Co-Expression Network Analysis and lncRNAs Annotation. Current Issues in Molecular Biology 2022; 44(4): 1479-1487. doi:10.3390/cimb44040100]